Resources developed in our lab
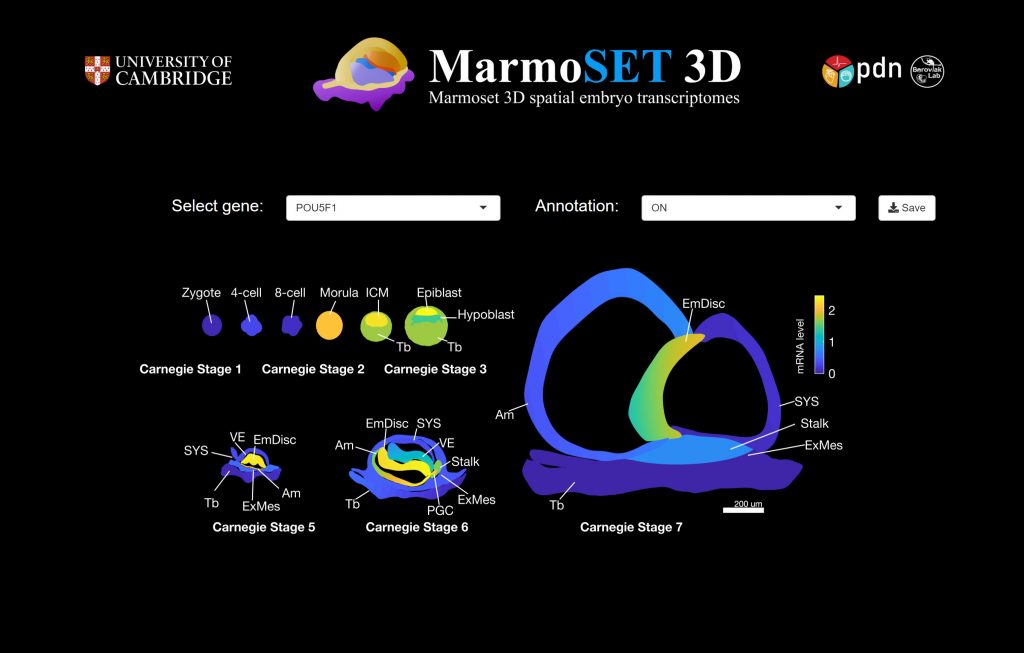
Marmoset 3D spatial embryo transcriptomes (MarmoSET 3D)
Our transcriptional blueprint of primate development from zygote to gastrulation (Nature 2022). Please allow 5-10 seconds to load in the spatial transcriptome dataset.
http://131.111.33.80:3838/marmoset3D/
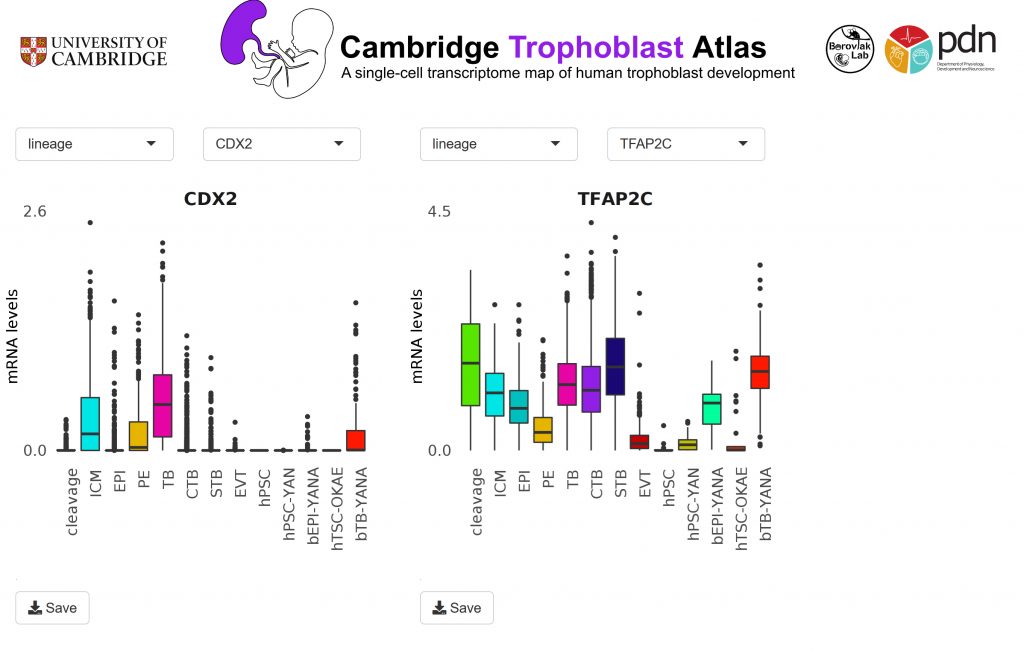
Cambridge Trophoblast Atlas
A single-cell transcriptome map of human trophoblast development developed from zygote to mid-gestation. Developed by the amazing Yutong Chen!
http://131.111.33.80:3838/TBatlas/
Genome-wide Rodent And Primate Preimplantation Atlas (GRAPPA)
Honouring its Italian creator, Giuliano Stirparo, in the name – GRAPPA is a fabulous tool to directly compare any gene of interest in human, marmoset and mouse preimplantation development.
https://app.stemcells.cam.ac.uk/GRAPPA/
Atlas of Human Preimplantation Embryo and In Vitro Cultured Cells
Here we present a direct comparison of human preimplantation development with a large selection of naive and primed pluripotent human emrbyonic stem cells lines in various culture conditions.
http://app.stemcells.cam.ac.uk/human-embryo/
Mouse and Marmoset Pathway Expression Atlas for Early Development
This online resource allows easy pairwise comparison of 294 signalling pathways between ESC, marmoset ICMs and various mouse preimplantation developmental stages. It also contains datasets of diapaused epiblasts, which can be compared to normal development, primates and ESC.
http://pathway-atlas.stemcells.cam.ac.uk